Description of the enrichment tests
Several target enrichment tests were benchmarked (results in the
benchmark tab), and are described below. The best overall tests were
selected as default for the app, and although other implemented tests are
made available in the app's advanced options, their usage is not
recommended.
The tests differ in the inputs they use, both in terms of the
target annotation as well as of the type of signal in which enrichment is
looked for. On the signal side, tests denoted as 'binary' compare features
(genes or transcripts) in a given set (e.g. your significantly downregulated
genes) to those in a background set (i.e. over-representation analysis),
whereas tests denoted as 'continuous' instead rely on a numeric input
signal, such as the mangitude or significance of changes in an input
differential expression analysis (by default, the tests use the sign of the
foldchange multiplied by the -log10(FDR), which is well correlated to logFC
for genes with low intra-group variability, and more robust than the latter).
On the annotation side, tests can also either use set membership (i.e.
whether or not a given feature is a predicted miRNA target) or numeric
values, such as the number of binding sites harbored by a given feature,
or a repression score (i.e. the extent to which a given feature is
predicted to be repressed by a miRNA).
As is the case for over-representation analysis in general, for
tests based on binary inputs the choice of a good background is
critical. In many contexts, the background set of genes will be the
set of genes expressed in the system of interest.
-
Default tests
-
siteoverlap
(binary signal, set membership):
The siteoverlap test is based on Fisher's
exact test, but using the number of sites on predicted
targets and in the background instead of counting each
feature as one. While in theory this violates the
assumption of independence of the counts (since all the
binding sites of a given transcript are either in or out
of the set), leading to slightly anti-conservative
p-values, in practice this test is excellent at
identifying the most enriched miRNA.
-
woverlap
(binary signal, set membership):
This test is like the above 'siteoverlap' test,
but corrects for UTR length using the Wallenius method, as implemented in
the
goseq
package. The test performs similarly to the siteoverlap test.
Note that it requires a certain number of sets with overlaps to be run.
-
areamir
(continuous signal, score or set
membership):
The areamir test is based on the analytic Rank-based
Enrichment Analysis (aREA) test implemented in the
'msviper' function of the
viper
package. The test is akin to an analytical version of
GSEA (see below), but it can additionally use degrees or
likelihood of set membership. If repression scores are
available in the annotation, areamir will therefore use a
(trimmed) version of it as set membership likelihood.
-
Other tests implemented and evaluated
-
overlap
(binary signal, set membership):
This test is based on Fisher's exact test, using the
number of features (i.e. transcripts/genes) among predicted targets
vs in the background (and therefore ignoring any site-based
information).
-
Mann-Whitney (MW)
(continuous signal, set membership):
This is the Mann-Whitney (also known as Wilcoxon)
non-parametric test comparing targets and non-targets. This
test performs badly in benchmarks and should not be used.
-
Kolmogorov-Smirnov (KS)
(continuous signal, set membership):
This is the Kolmogorov-Smirnov test comparing the
signal distribution of targets vs non-targets. This
test performs badly in benchmarks and should not be used.
-
modscore
(continuous signal, repression score):
This is a linear regression testing the relationship
between the input signal and the corresponding repression score
predicted for a given miRNA.
-
ebayes
(continuous signal, repression score):
This is akin to the `modscore` tests, but performed
using limma's moderated t-statistics.
-
lmadd
(continuous signal, repression score):
This is the `ebayes` tests, followed by consecutive
fits adding each top miRNAs to the previous ones in a single
model. This is especially useful to identify candidates which
are not redundant with the top hit.
-
modsites
(continuous signal, number of sites):
This is a linear regression testing the relationship
between the input signal and the number of predicted binding
sites for a given miRNA, correcting for UTR length.
-
GSEA
(continuous signal, set membership)
This test uses the multi-level fast GeneSet Enrichment
Analysis (GSEA) implemented in the
fgsea
package, which is highly successful for Gene Ontology enrichment
analysis. In the context of our benchmark, however, it performed very
poorly.
-
regmir
The regmir test uses constrained lasso-regularized regression,
which has a high specificity but lower sensitivity. The test will use
binary or continuous inputs (using then either linear or binomial
regression), as well as binary set membership or predicted repression
score, depending on the availability of the input. The binary version
of the test has shown the best performances.
Summary
Benchmark of the different target enrichment tests
The different tests were benchmarked on different datasets
each involving the transcriptomic characterization of the
knockdown or over-expression of different miRNAs. For each
experiment, the signal was additionally scrambled to create
further, more difficult 'pseudo-experiments', which are
averaged in the results below. The benchmark was performed
using TargetScan (conserved)-predicted sites, and was then
used to guide the choice of default tests in the enrichMiR app.
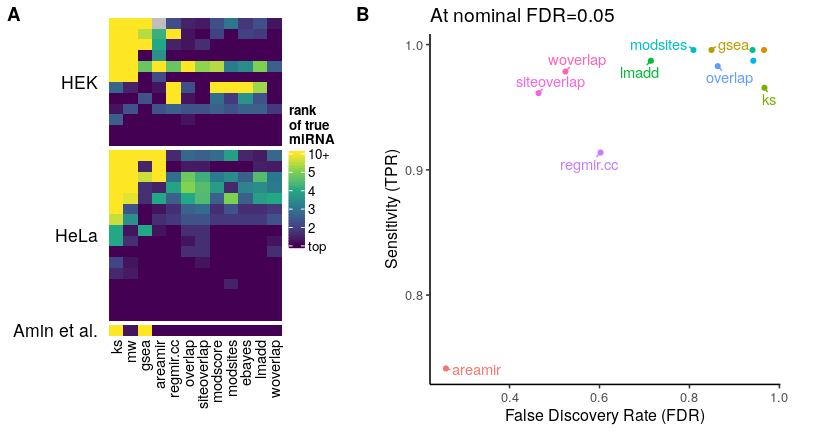
Panel A shows the rank of the true miRNA according to
the different tests (lower=better, i.e. a rank of 1
indicates that the true miRNA was correctly identified as
the top enriched one). Panel B shows the effective sensitivity
and False Discovery Rate (FDR) of the different tests at a
nominal q-value threshold of 0.05. One can observe that
while most tests manage to rank the true hypothesis as
first, most fail to accurately control error.
In light of these results, the siteoverlap and woverlap
tests were selected as the default for binary signals, and
the areamir test for continuous signals.
For
use with larger annotations (e.g. scanMiR), we however
recommend the more conservative lmadd test
(see
publication for details).
Note that restricting the enrichment analysis to the
miRNAs expressed in your system systematically decreases
FDR. You can do so in the 'Species and miRNAs' tab, either
using a custom list of miRNAs or selecting from available
tissues.

