Select a miRNA collection
Extra collection information
Display sites along transcript
Hover on points to view details, and click to visualize the alignment on the target sequence. You may also select miRNAs to show/hide by clicking on the legend.
Aggregated per miRNA
Table
Cached hits
Affinity plot
Targets
miRNA binding prediction
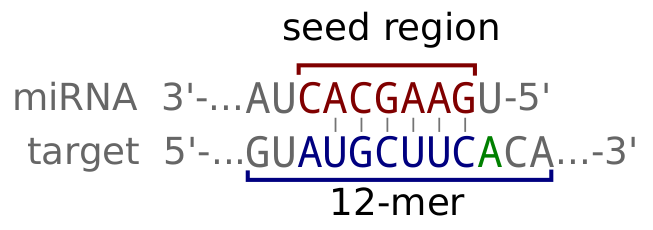
micro-RNAs (miRNAs) regulate transcripts stability and translation by targeting the Argonaute complex to RNAs exhibiting a partial complementarity with, in particular, the miRNA's seed sequence. For a general review on miRNA biology and targeting, see Bartel (2018). McGeary, Lin et al. (2019) have additionally shown the relevance of flanking nucleotides by empirically measuring the affinity (specifically the dissociation rate constant, KD) of a set of miRNAs to random 12-mer sequences, and then computationally extrapolating to other miRNAs). The scanMiR package, to which this app is an interface, builds on this work to offer a powerful and flexible binding and repression prediction framework.
Getting started
For a quick tour of the app, click here.
There are two main ways to use scanMiRApp:
Transcript-centered:
In the 'Search in gene/sequence' menu, you'll be able to scan the sequence of a given transcript for binding sites of (sets of) miRNA(s).
miRNA-centered:
In the 'miRNA-based' tab on the left, you'll be able to visualize information relative to a selected miRNA, including for instance its general binding profile and its top targets.
The shiny app was developed by Pierre-Luc Germain and
Michael Soutschek in the context of broader research in the Schratt lab.
Bugs reports and feature requests are welcome on the github repository.